TileDB-SOMA-ML
TileDB-SOMA-ML is a machine learning library for working with SOMA data formats.
See /shuffle for a visualization of the batching/shuffling algorithm:
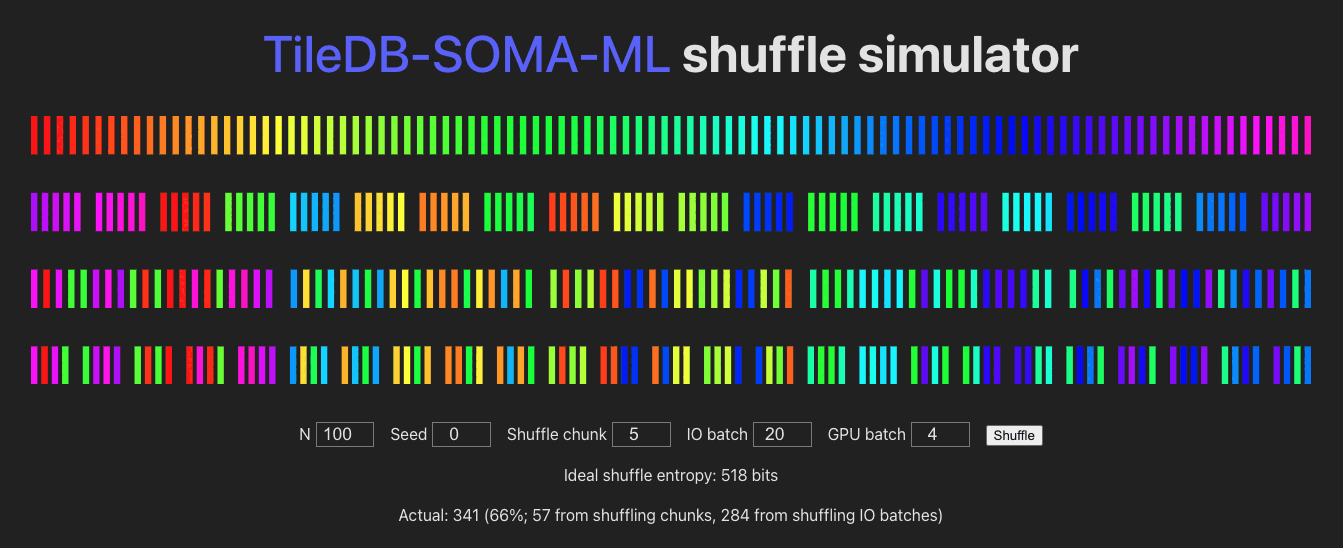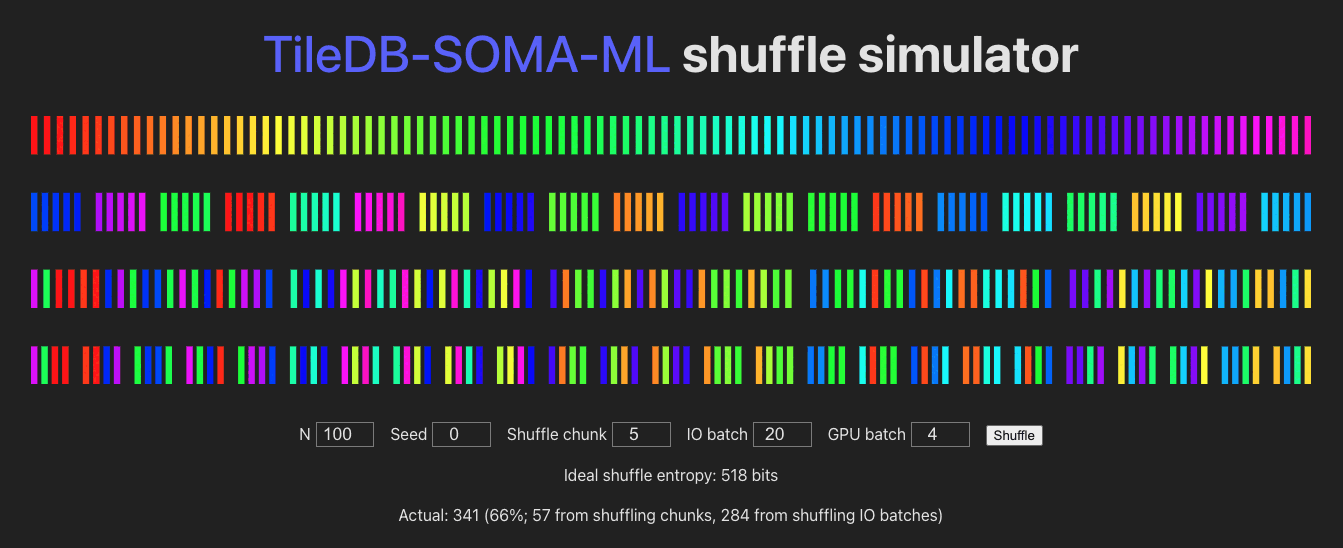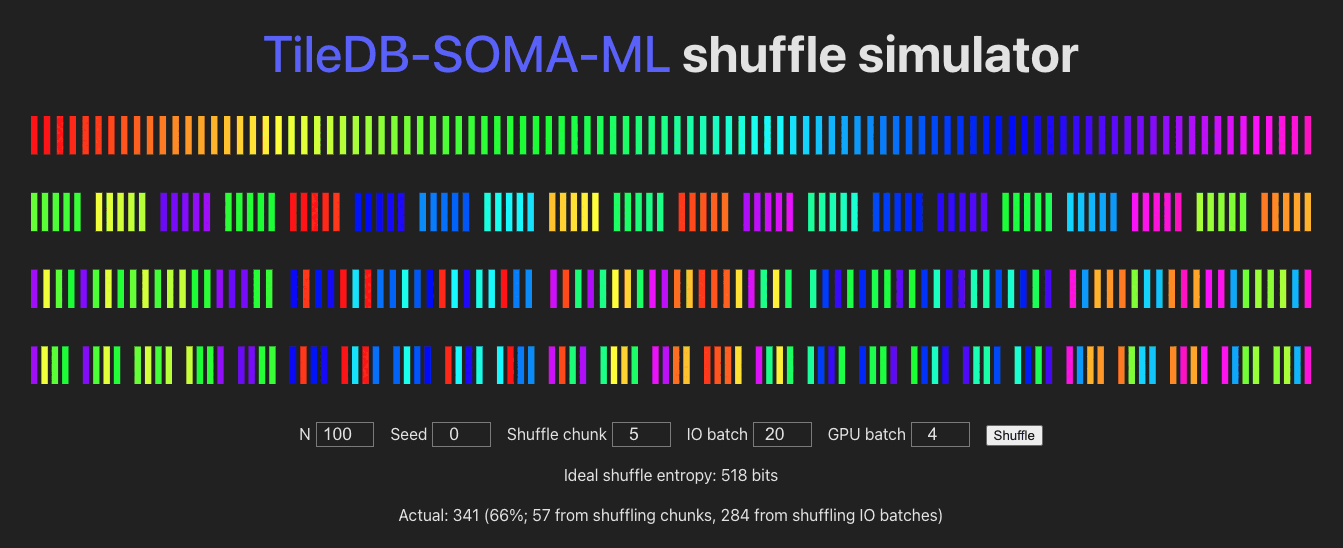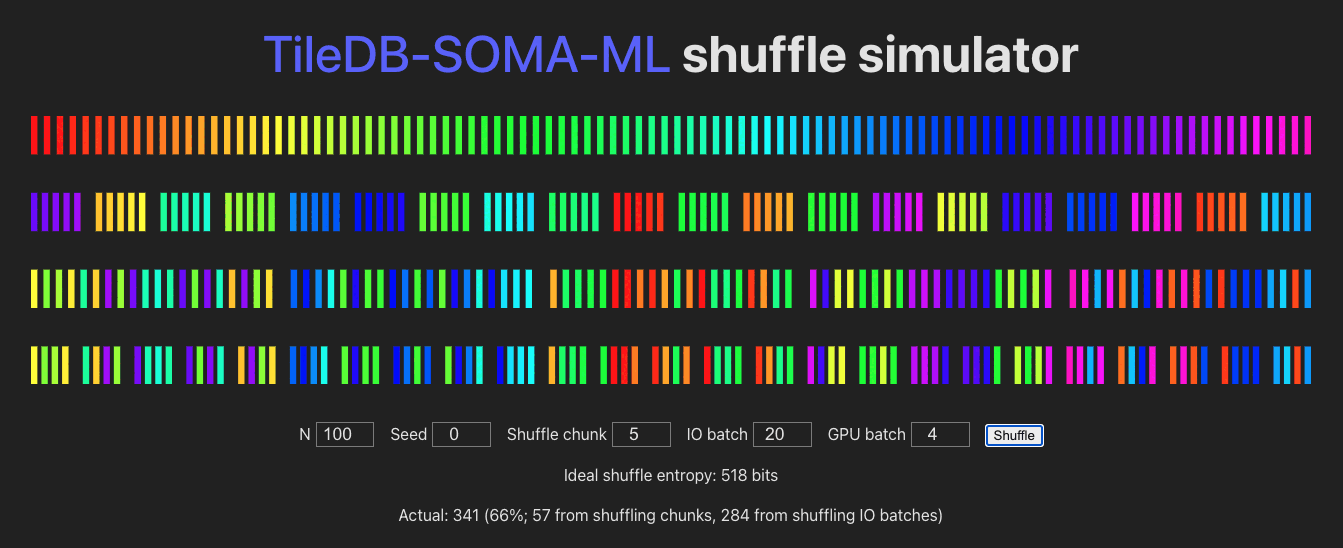
Module Contents
dataset
- class tiledbsoma_ml.ExperimentDataset(query: ExperimentAxisQuery | None = None, layer_name: str | None = None, x_locator: XLocator | None = None, query_ids: QueryIDs | None = None, obs_column_names: Sequence[str] = ('soma_joinid',), batch_size: int = 1, io_batch_size: int = 65536, shuffle: bool = True, shuffle_chunk_size: int = 64, seed: int | None = None, return_sparse_X: bool = False, use_eager_fetch: bool = True, shuffle_mode: ShuffleMode = ShuffleMode.CPU, device: device | None = None)[source]
An
IterableDatasetimplementation that reads from anExperimentAxisQuery.Provides an
IteratoroverMiniBatchs ofobsandXdata. EachMiniBatchis a tuple containing anndarrayand apd.DataFrame.An
ExperimentDatasetcan be passed toexperiment_dataloaderto enable multi-process reading/fetching.For example:
>>> from tiledbsoma import Experiment, AxisQuery >>> from tiledbsoma_ml import ExperimentDataset, experiment_dataloader >>> with Experiment.open("my_experiment_path") as exp: ... with exp.axis_query( ... measurement_name="RNA", ... obs_query=AxisQuery(value_filter="tissue_type=='lung'") ... ) as query: ... ds = ExperimentDataset(query) ... dl = experiment_dataloader(ds) >>> X_batch, obs_batch = next(iter(dl)) >>> X_batch array([0., 0., 0., ..., 0., 0., 0.], dtype=float32) >>> obs_batch soma_joinid 0 57905025
When
__iter__is invoked,obs_joinidsgoes through several partitioning, shuffling, and batching steps, ultimately yielding"mini batches"(tuples of matchedXandobsrows):Partitioning (
NDArrayJoinId):GPU-partitioning: if this is one of \(N>1\) GPU processes (see
get_distributed_rank_and_world_size),obs_joinidsis partitioned so that the $N$ GPUs will each receive the same number of samples (meaning up to $N-1$ samples may be dropped). Then, only the partition corresponding to the current GPU is kept, The resultingobs_joinidsis used in subsequent steps.DataLoader-worker partitioning: if this is one of $M>1$DataLoader-worker processes (seeget_worker_id_and_num),obs_joinidsis further split $M$ ways, and onlyobs_joinidscorresponding to the current process are kept.
Shuffle-chunking (
List[NDArrayJoinId]): ifshuffle=True,obs_joinidsare broken into “shuffle chunks”. The chunks are then shuffled amongst themselves (but retain their chunk-internal order, at this stage). Ifshuffle=False, one “chunk” is emitted containing allobs_joinids.IO-batching (
Iterable[IOBatch]): shuffle-chunks are re-grouped into “IO batches” of sizeio_batch_size. Ifshuffle=True, eachIOBatchis shuffled, then the correspondingXandobsrows are fetched from the underlyingExperiment.Mini-batching (
Iterable[MiniBatch]):IOBatchtuples are re-grouped into “mini batches” of sizebatch_size.
Shuffling support (in steps 2. and 3.) is enabled with the
shuffleparameter, and should be used in lieu ofDataLoader’s default shuffling functionality. Similarly,batch_sizeshould be used instead ofDataLoader’s default batching.experiment_dataloaderis the recommended way to wrap anExperimentDatasetin aDataLoader, as it enforces these constraints while passing through otherDataLoaderargs.Describing the whole process another way, we read randomly selected groups of
obscoordinates from across allExperimentAxisQueryresults, concatenate those into an I/O buffer, shuffle the buffer element-wise, fetch the full row data (Xandobs) for each coordinate, and send that on to PyTorch / the GPU, in mini-batches. The randomness of the shuffle is determined by:shuffle_chunk_size: controls the granularity of the global shuffle.shuffle_chunk_size=1corresponds to a full global shuffle, but decreases I/O performance. Larger values cause chunks of rows to be shuffled, increasing I/O performance (by taking advantage of data locality in the underlyingExperiment) but decreasing overall randomness of the yielded data.io_batch_size: number of rows to fetch at once (comprised of concatenated shuffle-chunks, and shuffled row-wise). Larger values increase shuffle-randomness (by shuffling more “shuffle chunks” together), I/O performance, and memory usage.
- Lifecycle:
experimental
- __init__(query: ExperimentAxisQuery | None = None, layer_name: str | None = None, x_locator: XLocator | None = None, query_ids: QueryIDs | None = None, obs_column_names: Sequence[str] = ('soma_joinid',), batch_size: int = 1, io_batch_size: int = 65536, shuffle: bool = True, shuffle_chunk_size: int = 64, seed: int | None = None, return_sparse_X: bool = False, use_eager_fetch: bool = True, shuffle_mode: ShuffleMode = ShuffleMode.CPU, device: device | None = None)[source]
Construct a new
ExperimentDataset.- Parameters:
query –
ExperimentAxisQuerydefining data to iterate over. This constructor requires {query,layer_name} xor {x_locator,query_ids}.layer_name –
Xlayer to read. This constructor requires {query,layer_name} xor {x_locator,query_ids}.x_locator –
XLocatorpointing to anXarray to read. This constructor requires {query,layer_name} xor {x_locator,query_ids}.query_ids –
QueryIDscontainingobsandvarjoinids to read. This constructor requires {query,layer_name} xor {x_locator,query_ids}.obs_column_names – The names of the
obscolumns to return. At least one column name must be specified. Default is('soma_joinid',).batch_size – The number of rows of
Xandobsdata to yield in eachMiniBatch. Whenbatch_sizeis 1 (the default) andreturn_sparse_XisFalse(also default), the yieldedndarrays will have rank 1 (representing a single row); larger values ofbatch_size(orreturn_sparse_XisTrue) will result in arrays of rank 2 (multiple rows). Note that abatch_sizeof 1 allows thisIterableDatasetto be used withDataLoaderbatching, but higher performance can be achieved by performing batching in this class, and setting theDataLoadersbatch_sizeparameter toNone.io_batch_size –
The number of
obs/Xrows to retrieve when reading data from SOMA. This impacts:Maximum memory utilization, larger values provide better read performance, but require more memory.
The number of rows read prior to shuffling (see the
shuffleparameter for details).
The default value of 65,536 provides high performance but may need to be reduced in memory-limited hosts or when using a large number of
DataLoaderworkers.shuffle – Whether to shuffle the
obsandXdata being returned. Defaults toTrue.shuffle_chunk_size – Global-shuffle granularity; larger numbers correspond to less randomness, but greater read performance. “Shuffle chunks” are contiguous rows in the underlying
Experiment, and are shuffled among themselves before being combined into IO batches (which are internally shuffled, before fetching and finally mini-batching). Ifshuffle == False, this parameter is ignored.seed – The random seed used for shuffling. Defaults to
None(no seed). This argument MUST be specified when usingDistributedDataParallelto ensure data partitions are disjoint across worker processes.return_sparse_X – If
True, will return theXdata as acsr_matrix. IfFalse(the default), will returnXdata as andarray.use_eager_fetch – Fetch the next SOMA chunk of
obsandXdata immediately after a previously fetched SOMA chunk is made available for processing via the iterator. This allows network (or filesystem) requests to be made in parallel with client-side processing of the SOMA data, potentially improving overall performance at the cost of doubling memory utilization. Defaults toTrue.
- Raises:
ValueError – on unsupported or malformed parameter values.
- Lifecycle:
experimental
Warning
When using this class in any distributed mode, calling the
set_epoch()method at the beginning of each epoch before creating theDataLoaderiterator is necessary to make shuffling work properly across multiple epochs. Otherwise, the same ordering will always be used.In addition, when using shuffling in a distributed configuration (e.g.,
DDP), you must provide a seed, ensuring that the same shuffle is used across all replicas.
- __iter__() Iterator[Tuple[ndarray[tuple[Any, ...], dtype[number[Any]]] | csr_matrix, DataFrame]][source]
Emit
MiniBatchs (alignedXandobsrows).- Lifecycle:
experimental
- __len__() int[source]
Return the number of batches this iterable will produce. If run in the context of
torch.distributedor as a multi-process loader (i.e.,DataLoaderinstantiated with num_workers > 0), the batch count will reflect the size of the data partition assigned to the active process.See important caveats in the PyTorch
DataLoaderdocumentation regardinglen(dataloader), which also apply to this class.- Returns:
int(number of batches).
- Lifecycle:
experimental
- io_batch_size: int
Number of
obs/Xrows to fetch together, when reading from the providedExperimentAxisQuery.
- query_ids: QueryIDs
obs/varcoordinates (from anExperimentAxisQuery) to iterate over.
- random_split(*fracs: float, seed: int | None = None, method: Literal['deterministic', 'multinomial', 'stochastic_rounding'] = 'stochastic_rounding') Tuple[ExperimentDataset, ...][source]
Split this
ExperimentDatasetinto 1 or moreExperimentDataset‘s, randomly sampled accordingfracs.fracsmust sum to $1$seedis optionalmethod: seeSamplingMethodfor details
- return_sparse_X: bool
When
True, returnXdata as acsr_matrix(by default, returnndarrays).
- set_epoch(epoch: int) None[source]
Set the epoch for this Data iterator.
When
shuffleisTrue, this will ensure that all replicas use a different random ordering for each epoch. Failure to call this method before each epoch will result in the same data ordering across all epochs.This call must be made before the per-epoch iterator is created.
- property shape: Tuple[int, int]
Return the number of batches and features that will be yielded from this
Experiment.If used in multiprocessing mode (i.e.
DataLoaderinstantiated with num_workers > 0), the number of batches will reflect the size of the data partition assigned to the active process.- Returns:
number of batches, number of vars.
- Return type:
A tuple of two
intvalues
- Lifecycle:
experimental
- shuffle_chunk_size: int
Number of contiguous rows shuffled as an atomic unit (before later concatenation and shuffling within
IOBatchs).
- shuffle_mode: ShuffleMode
Whether to shuffle on cpu or gpu (and at what granularity).
Only read when shuffle=True
- x_locator: XLocator
State required to open an
XSparseNDArray(and associatedobsDataFrame), within anExperiment.
dataloader
- tiledbsoma_ml.experiment_dataloader(ds: ExperimentDataset, **dataloader_kwargs: Any) DataLoader[source]
DataLoaderfactory method for safely wrapping anExperimentDataset.Several
DataLoaderconstructor parameters are not applicable, or are non-performant when using loaders from this module, includingshuffle,batch_size,sampler, andbatch_sampler. Specifying any of these parameters will result in an error.Refer to the DataLoader docs for more information on
DataLoaderparameters, andExperimentDatasetfor info on corresponding parameters.- Parameters:
ds – A
IterableDataset. May include chained data pipes.**dataloader_kwargs – Additional keyword arguments to pass to the
DataLoaderconstructor, except forshuffle,batch_size,sampler, andbatch_sampler, which are not supported when using data loaders in this module.
- Returns:
- Raises:
ValueError – if any of the
shuffle,batch_size,sampler, orbatch_samplerparams are passed as keyword arguments.
- Lifecycle:
experimental
scvi
- tiledbsoma_ml.SCVIDataModule(query: ExperimentAxisQuery, *args: Any, batch_column_names: Sequence[str] | None = None, batch_labels: Sequence[str] | None = None, dataloader_kwargs: dict[str, Any] | None = None, train_size: float = 1.0, seed: int = 42, **kwargs: Any)[source]
PyTorch Lightning DataModule for training scVI models from SOMA data.
Wraps a
ExperimentDatasetto stream the results of a SOMAExperimentAxisQuery, exposing aDataLoaderto generate tensors ready for scVI model training. Also handles deriving the scVI batch label as a tuple of obs columns.- Lifecycle:
Experimental.
Batching and Data Management
_common
Type aliases used in ExperimentDataset and experiment_dataloader.
- tiledbsoma_ml._common.MiniBatch
Yielded by
ExperimentDataset; pairs a slice ofXrows with correspondingobsrows.When
return_sparse_XisFalse(the default), aMiniBatchis a tuple ofndarrayandpd.DataFrame(forXandobs, respectively). Ifbatch_size=1, thendarraywill have rank 1 (representing a single row), otherwise it will have rank 2. Ifreturn_sparse_XisTrue, theXslice is returned as acsr_matrix(which is always rank 2).alias of
Tuple[ndarray[tuple[Any, …],dtype[number[Any]]] |csr_matrix,DataFrame]
_query_ids
Shuffle-chunk and partition (across GPU and DataLoader-worker processes) while reading from a SOMA Experiment.
QueryIDs
- class tiledbsoma_ml._query_ids.QueryIDs(*, obs_joinids: ndarray[tuple[Any, ...], dtype[int64]], var_joinids: ndarray[tuple[Any, ...], dtype[int64]], partition: Partition | None = None)[source]
Wrapper for obs and var IDs from an
ExperimentAxisQuery.Serializable across multiple processes.
- partition: Partition | None
GPU/Worker-partition info; typically populated by
partitioned()
- classmethod create(query: ExperimentAxisQuery) QueryIDs[source]
Initialize a
QueryIDsobject from anExperimentAxisQuery.
- random_split(*fracs: float, seed: int | None = None, method: Literal['deterministic', 'multinomial', 'stochastic_rounding'] = 'stochastic_rounding') Tuple[QueryIDs, ...][source]
Split this
QueryIDsinto 1 or moreQueryIDs, randomly sampled accordingfracs.fracsmust sum to $1$seedis optionalmethod: seeSamplingMethodfor details
- partitioned(partition: Partition) QueryIDs[source]
Create a new
QueryIDswithobs_joinidscorresponding to a given GPU/workerPartition.If
Noneis provided, world size, rank, num workers, and worker ID will be inferred using helper functions that read env vars (seeget_distributed_rank_and_world_size,get_worker_id_and_num).When
WORLD_SIZE > 1, each GPU will receive the same number of samples (meaning up toWORLD_SIZE-1samples may be dropped).
- shuffle_chunks(shuffle_chunk_size: int, seed: int | None = None) List[ndarray[tuple[Any, ...], dtype[int64]]][source]
Divide
obs_joinidsinto chunks of sizeshuffle_chunk_size, and shuffle them.Used as a compromise between a full random shuffle (optimal for training performance/convergence) and a sequential, un-shuffled traversal (optimal for I/O efficiency).
Partition
Chunks
- tiledbsoma_ml._query_ids.Chunks
Return-type of
QueryIDs.shuffle_chunks,Listofndarrays.
SamplingMethod
- tiledbsoma_ml._query_ids.SamplingMethod
Enum arg to
QueryIDs.random_split:"deterministic": number of each class returned will always be \(frac \times N\), rounded to nearest int, e.g.n=12, fracs=[.7,.3]will always produce 8 and 4 elements, resp."multinomial": each element is assigned to a class independently; no guarantees are made about resulting class sizes."stochastic_rounding": guarantee each class gets assigned at least \(\lfloor frac \times N \rfloor\) elements. The remainder are then distributed so that class-size expected-values match the providedfracs.
_io_batch_iterable
- tiledbsoma_ml._io_batch_iterable.IOBatch
Tuple type emitted by
IOBatchIterable, containingXrows (as aCSR_IO_Buffer) andobsrows (pd.DataFrame).alias of
Tuple[CSR_IO_Buffer,DataFrame]
- class tiledbsoma_ml._io_batch_iterable.IOBatchIterable(chunks: List[ndarray[tuple[Any, ...], dtype[int64]]], io_batch_size: int, obs: DataFrame, var_joinids: ndarray[tuple[Any, ...], dtype[int64]], X: SparseNDArray, obs_column_names: Sequence[str] = ('soma_joinid',), seed: int | None = None, shuffle: bool = True, use_eager_fetch: bool = True)[source]
Given a list of
obs_joinidChunks, re-chunk them into (optionally shuffled)IOBatch’s”.An
IOBatchis a tuple consisting of a batch of rows from theXSparseNDArray, as well as the corresponding rows from theobsDataFrame. TheXrows are returned in an optimizedCSR_IO_Buffer.
_mini_batch_iterable
- class tiledbsoma_ml._mini_batch_iterable.MiniBatchIterable(io_batch_iter: IOBatchIterable, batch_size: int, use_eager_fetch: bool = True, return_sparse_X: bool = False, gpu_shuffle: bool = False, gpu_shuffle_mode: str = 'iobatch', device: device | None = None, seed: int | None = None, epoch: int = 0)[source]
Convert (possibly shuffled)
IOBatchIterableintoMiniBatch’s suitable for passing to PyTorch.- io_batch_iter: IOBatchIterable
x_locator
- class tiledbsoma_ml.x_locator.XLocator(*, uri: str, measurement_name: str, layer_name: str, tiledb_timestamp_ms: int, tiledb_config: Dict[str, str | float])[source]
State required to open an
XSparseNDArray(and associatedobsDataFrame), within anExperiment.Serializable across multiple processes.
- classmethod create(experiment: Experiment, measurement_name: str, layer_name: str) XLocator[source]
Initialize an
XLocatorobject from anExperiment,measurement_name, andlayer_name.The arguments provide sufficient info to identify a specific
X“layer” in the providedExperiment.
_distributed
Utilities for multiprocess training: determine GPU “rank” / “world_size” and DataLoader worker ID / count.
- tiledbsoma_ml._distributed.get_distributed_rank_and_world_size() Tuple[int, int][source]
Return tuple containing equivalent of
torch.distributedrank and world size.
- tiledbsoma_ml._distributed.get_worker_id_and_num() Tuple[int, int][source]
Return
DataLoaderID, and the total number ofDataLoaderworkers.
- tiledbsoma_ml._distributed.init_multiprocessing() None[source]
Ensures use of “spawn” for starting child processes with multiprocessing.
Note
Forked processes are known to be problematic: Avoiding and fighting deadlocks.
CUDA does not support forked child processes: CUDA in multiprocessing.
Private.
_csr
CSR sparse matrix implementation, optimized for incrementally building from COO matrices.
Private module.
- class tiledbsoma_ml._csr.CSR_IO_Buffer(indptr: ndarray[tuple[Any, ...], dtype[unsignedinteger[Any]]], indices: ndarray[tuple[Any, ...], dtype[unsignedinteger[Any]]], data: ndarray[tuple[Any, ...], dtype[number[Any]]], shape: Tuple[int, int])[source]
Implement a minimal CSR matrix with specific optimizations for use in this package.
- Operations supported are:
Incrementally build a CSR from COO, allowing overlapped I/O and CSR conversion for I/O batches, and a final “merge” step which combines the result.
Zero intermediate copy conversion of an arbitrary row slice to dense (i.e., mini-batch extraction).
Parallel processing, where possible (construction, merge, etc.).
Minimize memory use for index arrays.
Overall is significantly faster, and uses less memory, than the equivalent
scipy.sparseoperations.- __init__(indptr: ndarray[tuple[Any, ...], dtype[unsignedinteger[Any]]], indices: ndarray[tuple[Any, ...], dtype[unsignedinteger[Any]]], data: ndarray[tuple[Any, ...], dtype[number[Any]]], shape: Tuple[int, int]) None[source]
Construct from PJV format.
- static from_ijd(i: ndarray[tuple[Any, ...], dtype[unsignedinteger[Any]]], j: ndarray[tuple[Any, ...], dtype[unsignedinteger[Any]]], d: ndarray[tuple[Any, ...], dtype[number[Any]]], shape: Tuple[int, int]) CSR_IO_Buffer[source]
Build a
CSR_IO_Bufferfrom a COO sparse matrix representation.
- static from_pjd(p: ndarray[tuple[Any, ...], dtype[unsignedinteger[Any]]], j: ndarray[tuple[Any, ...], dtype[unsignedinteger[Any]]], d: ndarray[tuple[Any, ...], dtype[number[Any]]], shape: Tuple[int, int]) CSR_IO_Buffer[source]
Build a
CSR_IO_Bufferfrom a SCR sparse matrix representation.
- property dtype: type[Any] | dtype[Any] | _SupportsDType[dtype[Any]] | tuple[Any, Any] | list[Any] | _DTypeDict | str | None
Underlying Numpy dtype.
- slice_tonumpy(row_index: slice) ndarray[tuple[Any, ...], dtype[number[Any]]][source]
Extract slice as a dense ndarray.
Does not assume any particular ordering of minor axis.
- slice_toscipy(row_index: slice) csr_matrix[source]
Extract slice as a
sparse.csr_matrix.Does not assume any particular ordering of minor axis, but will return a canonically ordered scipy sparse object.
- static merge(mtxs: Sequence[CSR_IO_Buffer]) CSR_IO_Buffer[source]
Merge
CSR_IO_Buffers.